Note
Go to the end to download the full example code
Convert TRC file to mne format#
Here we convert a trc file recorded with a Micromed system to fif, the common format used in mne framework.
# Author: Etienne de Montalivet <etienne.demontalivet@protonmail.com>
#
# License: BSD-3-Clause
from micromed_io.to_mne import create_mne_from_micromed_recording
from pathlib import Path
fname = Path("../data/sample.TRC")
mne_raw = create_mne_from_micromed_recording(fname)
WARNING:root:MKR channel(s) is not close to 50mV. If no trigger received, then data are corrupted.
Creating RawArray with float64 data, n_channels=14, n_times=349632
Range : 0 ... 349631 = 0.000 ... 170.718 secs
Ready.
Info from trc file is parsed and stored in mne.Info
mne_raw.info
As you see, markers sent by serial connection to Micromed are parsed from the trc file and stored as mne.Annotations scalings: 7e-3 comes from Micromed +/- 3.2mV
mne_raw.plot(scalings=7e-3, duration=20, start=20)
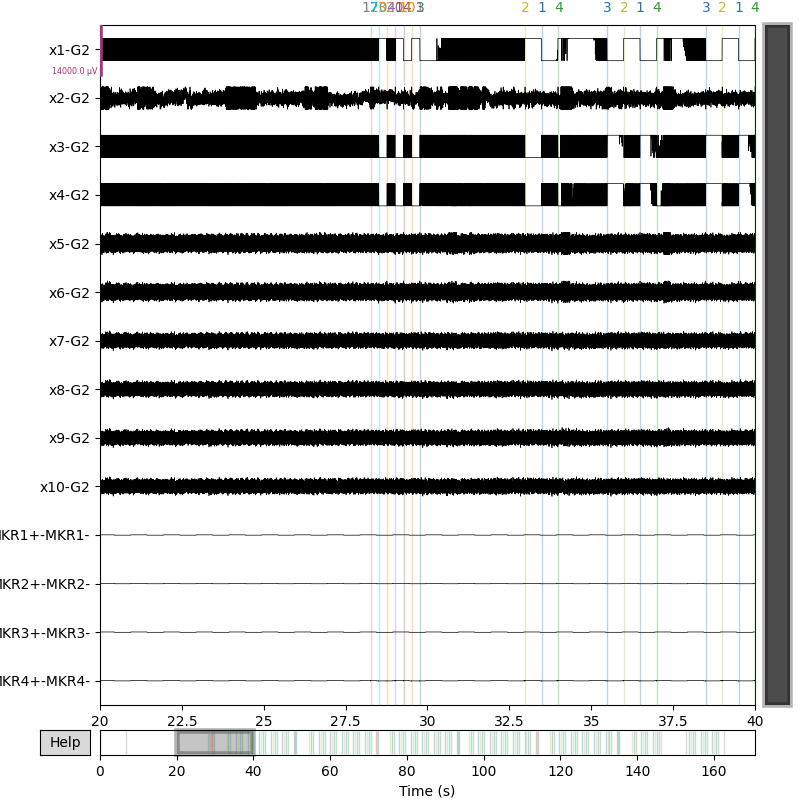
Using matplotlib as 2D backend.
<MNEBrowseFigure size 800x800 with 4 Axes>
Total running time of the script: (0 minutes 2.144 seconds)